DFAST Help
DDBJ Fast Annotation and Submission Tool (DFAST) is a bacterial genome annotation pipeline integrated with quality and taxonomy assessment methods. DFAST is developed so that all the procedure required for submission can be done seamlessly on-line, thus it can be used as an on-line workspace to prepare submission files to DDBJ Mass Submission System (MSS).
You can access the Job Submission Form from "Analysis > Genome Annotation" in the menu bar.
As of August 2017, we have replaced the background annotation engine of DFAST with a newly-implemented pipeline called DFAST-core.
Please note that the description shown in this page might be different from the current version, although the basic usage is the same.
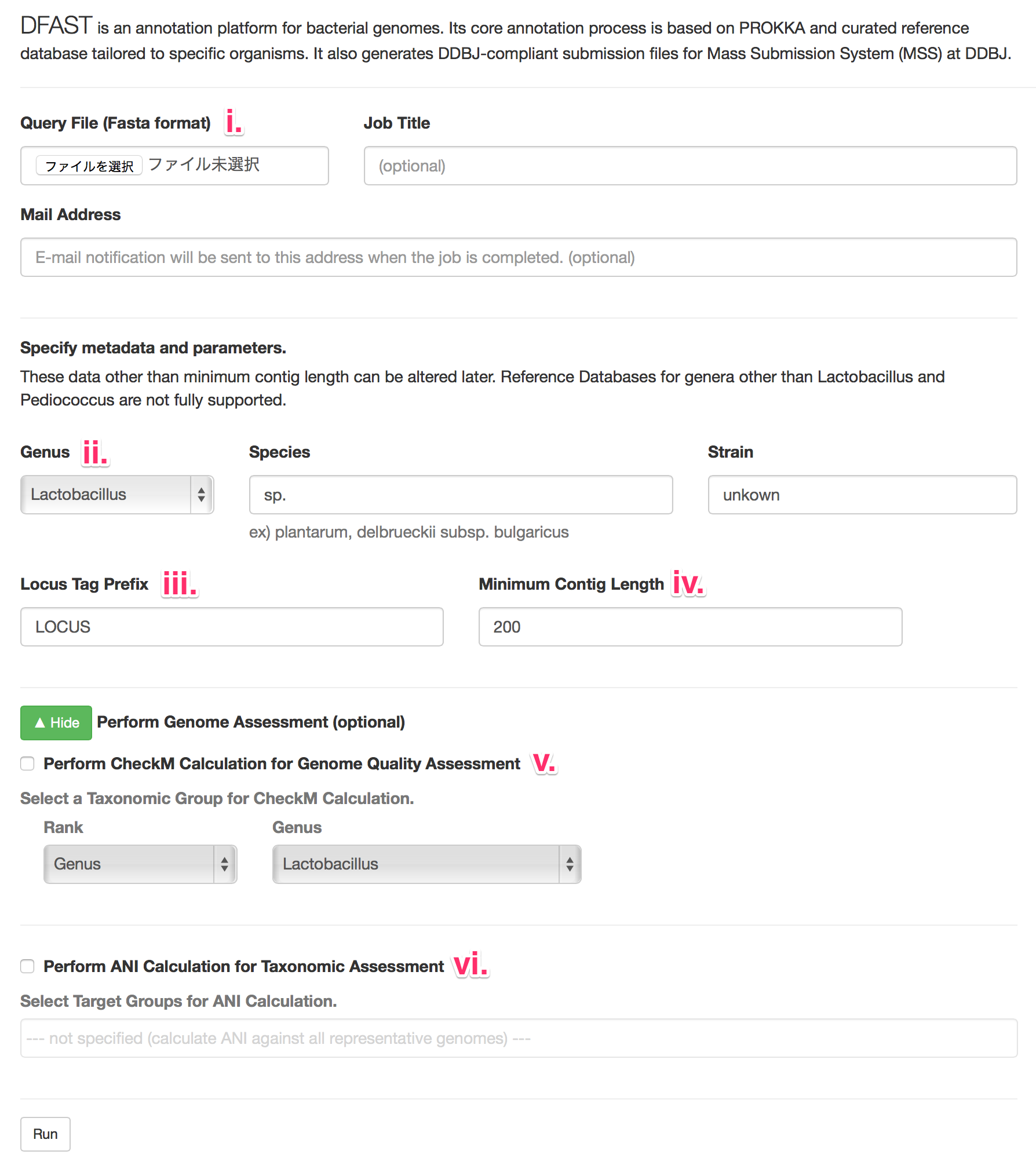
- i. Qury File
- Only fasta-formated file (<15Mbyte) is acceptable.
- Compressed files (.zip, .gz, or .bz2) are not acceptable.
- ii. Organism Name
- Genus, Species, and Strain are required, and can be moddified later.
- iii. Locus Tag Prefix
- Required, and this can be moddified later.
- Locus_tags are identifiers that are systematically applied to every gene in a genome. You need to register locus_tag prefix befor submitting the genome to INSDC.
- Please refer to the guideline of DDBJ for more information. Japanese / English
- iv. Minimum Contig Length
- Contigs shorter than this length will be eliminated.
- The default value of 200 bp is the recommendation of INSDC.
- Please refer to the NCBI WGS sumbission guideline for more information.
- v. Check here to perform Genome Quality Assessment using CheckM.
- [This service is currently only available for Lactic acid bacteria]
CheckM estimates genome completeness and contamination by inspecting the presence/absence of marker genes specific for a given taxon. Please refer to the original paper of CheckM for detailed description. - Normally, Rank and Genus are automatically specified, but you can also specify them manually.
- vi. Check here to perform Quality Assessment using ANI.
- [This service is currently only available for Lactic acid bacteria]
ANI (average nucleotide identity) represents the mean of sequence identity of homologous regions in the alignment between a given pair of genomes. A widely accepted threshold of the ANI value for distinguishing species is 95-96%. We followed the method described by Goris. et al. to calculate ANI. - This pipeline performs ANI calculation against representative genomes (mainly type strains) deposited in DAGA. If "Target Groups" are not specified, ANI calculation will be performed against all representative genomes (This which may take a while).
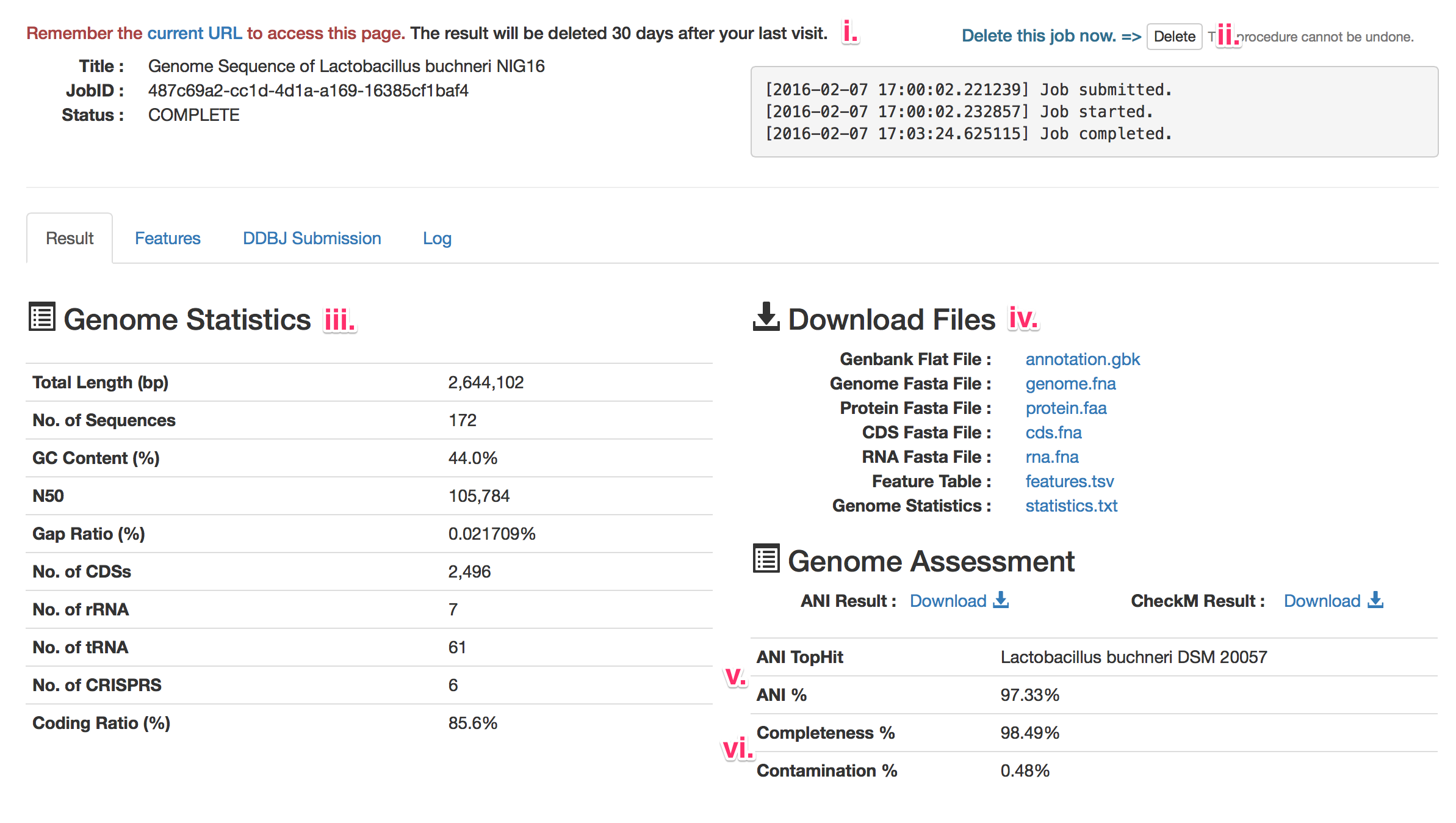
- i. Access to the annotation result.
- DFAST issues a uniq identifier to each job, which will be embedded in the URL for the result page. Be sure to remember the URL to access the result page again.
- The result will be automatically deleted 30 days after the last visit.
- ii. Delete the job.
- Click here to delete the job. This procedure cannot be undone.
- iii. Genome Statistics
- Several statistical metrics, such as N50 and number of coding sequences, are shown.
- iv. Download files.
- The annotation result can be downloaded in several formats.
- If you have editted the features, files will be updated accordingly.
- v. Result of Taxonomic Assessment.
- "ANI TopHit" shows the organism name that shared the highest ANI value with the query genome. If "ANI %" exceeds the threshold to discriminate species (around 95%), it is probable that the query geonome belongs to the same species as the one shown in "ANI TopHit".
- vi. Result of Genome Quality Assessment.
- Completeness is estimated by the number of single-copy gene markers identified in the genome, and Contamination is estimated from the multiplicity of the markers. Please refer to the original paper of CheckM for detailed description.
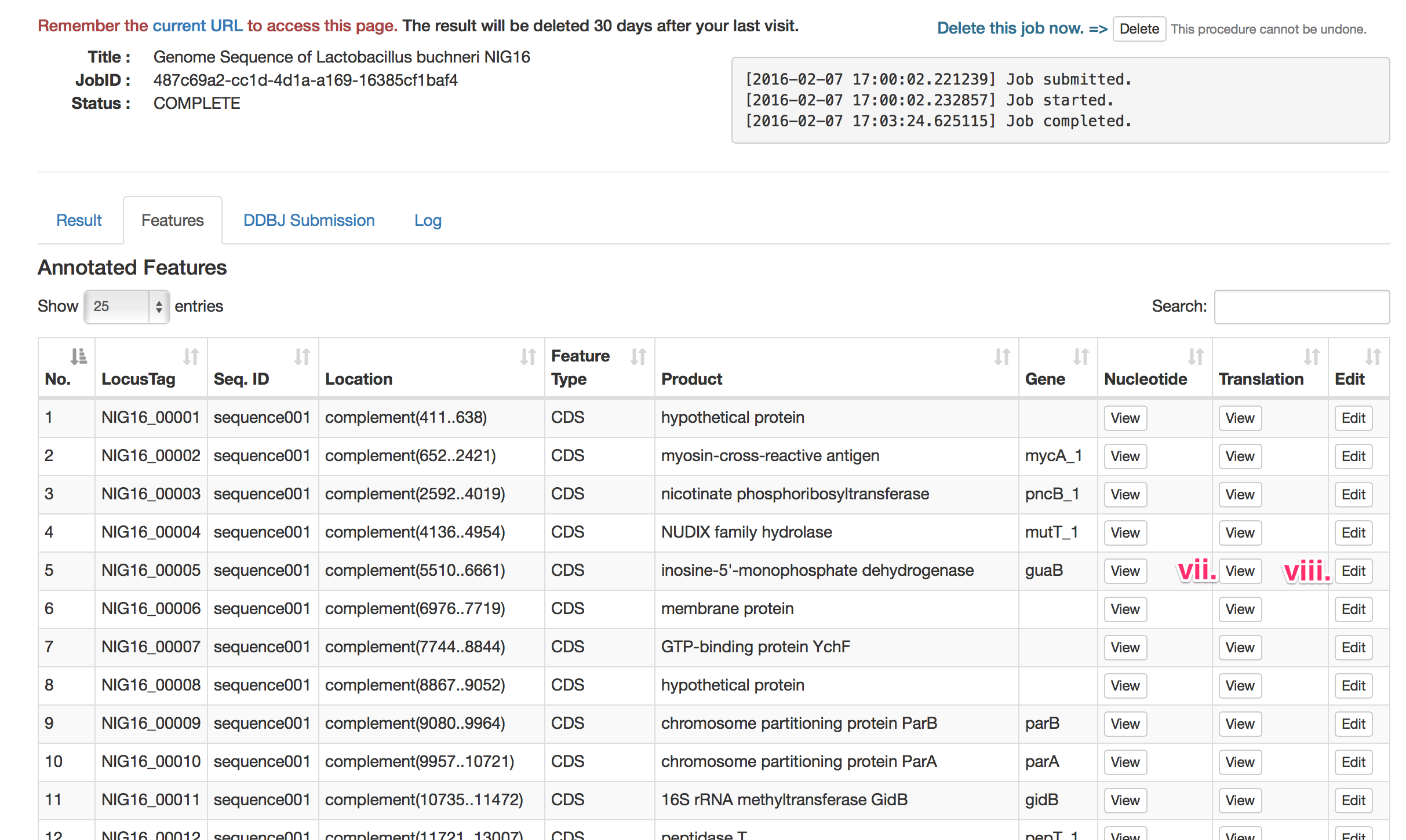
- vii. View Nucleotide or protein sequences.
- You can see the nucleotide or protein sequences. External link to NCBI BLAST service is also available.
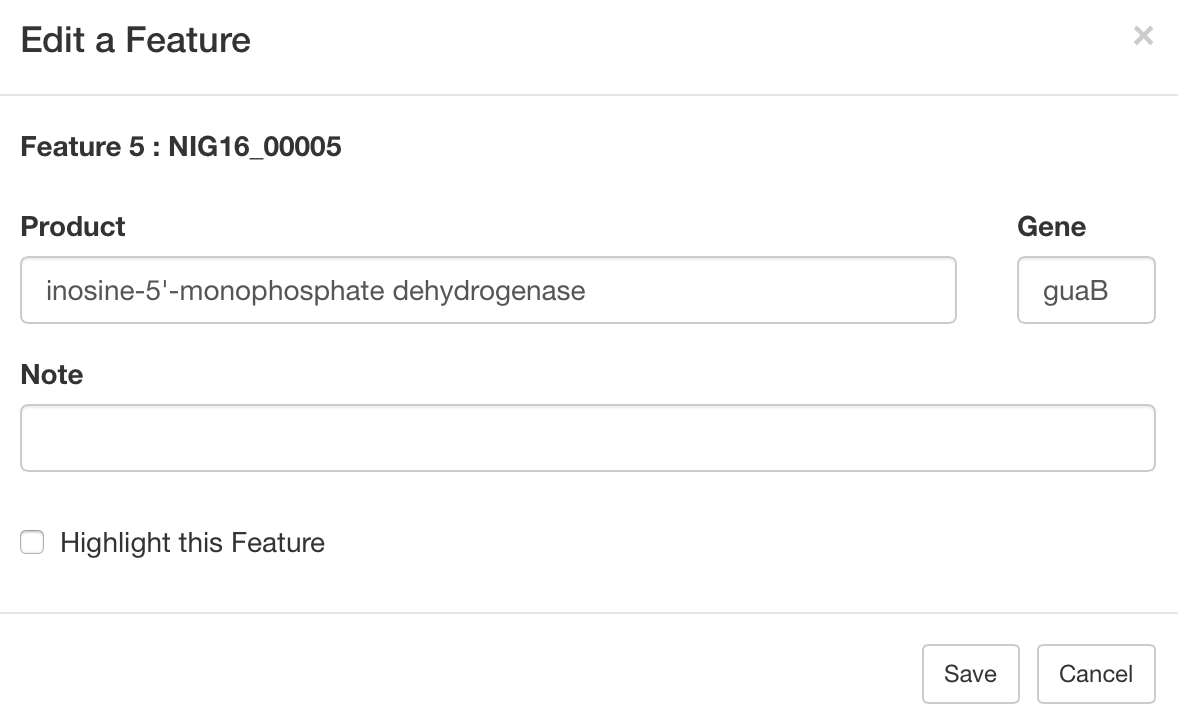
- viii. Edit the feature.
- You can edit the product name and gene symbol. You can also add the note.
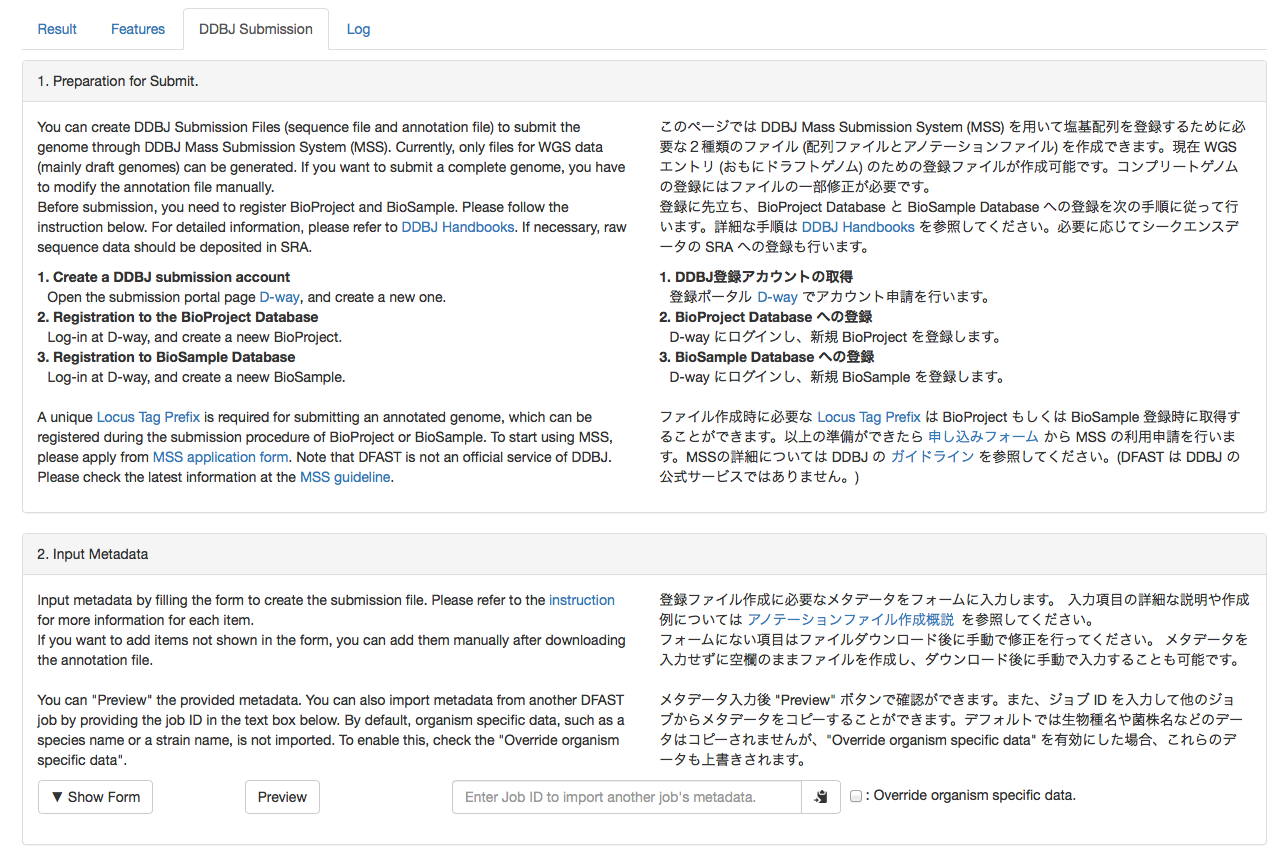
You can create submission files to DDBJ Mass Subission System (MSS) here. Please follow the instruction shown in the page to create files.
Please also refer to the following guidelines for the latest information.
DDBJ Submission Guideline:
Japanese /
English
DDBJ MSS:
Japanese /
English
DDBJ WGS Submission Guideline:
Japanese /
English